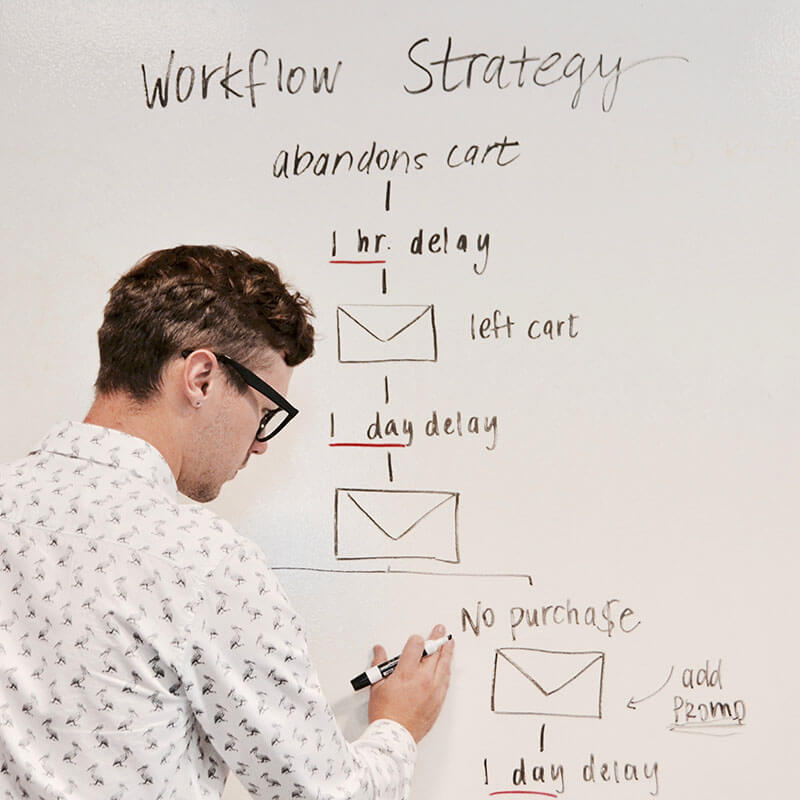
Mission statement
Genomic Ecology is an interdisciplinary research field at the interface between ecology and evolution, molecular biology, and genomics. It forges the links between molecular biologists who work with cutting-edge genomics (and ”post-genomic” tools),evolutionary biologists and ecologists who aim to understand the genetic mechanisms that underlie the process of evolution: changes in gene frequencies, adaptation, and interactions between organisms. In this “spirit” GENECO should Provide a platform for research collaboration and exchange of techniques, theory, & study systems in genomic ecology Train PhD students at the highest international scientific standard Create a doctoral education that will prepare for a career both within and outside academia
Courses
GENECO courses are generally one week long and are given at the Biology Department at Lund University. The one-week courses are free of charge and open to PhD students from any university, although GENECO students are given priority. The curriculum offers both introductory and specialized courses, as well as hands-on laboratory (wet or computer) and theoretical courses.
Workshops are open to all participants (PhD students, postdocs, PhDs) and are of different lengths. They are usually not held in Lund.
Courses Fall 2010 - Winter 2011
28/06/2010
Evolutionary Quantitative Genetics (Start October 14, 1 meeting/week)*
AFLP for Population Genetics (November 8-12)*
Conservation Genetics in a changing world (November 22-24)*
Fermentors, HPLC and Experimental Evolution (December 6-10)*
Comparative Genomics (January 9-21)
Intro Course: Molecular Ecology and Evolution (January 18-February 4)
Workshop on Comparative Genomics
17/08/2010
The Workshop on Comparative Genomics consists of a series of lectures, demonstrations and computer laboratories that cover various aspects of comparative genomics. Faculty are chosen exclusively for their effectiveness in teaching theory and practice in comparative genomics. Included among the faculty are developers and other experts in the use of computer programs and packages such as Ensembl and Galaxy who provide demonstrations and consultations. The course is designed for established investigators, postdoctoral scholars, and advanced graduate students. Scientists with strong interests in the uses of short-read sequence data, analytical methods, comparative structure of genomes, SNP detection and analysis, CNV, genome visualization tools and related areas are encouraged to apply for admission. Lectures and computer laboratories total ~90 hours of scheduled instruction. Admission is limited and highly competitive, with admissions decisions determined by an international committee.
Topics to be covered include: Sequencing technologies: short-read sequencing technologies of various types Assembly and alignment: basic analyses in de novo and resequencing studies Gene finding and annotation: functional description of genomic data Genome characterization: gene content; genome structure; synteny; SNPs; copy number variation (CNV) Assigning sequences to taxonomic groups in metagenomic studies: moving from sequences of unknown taxa to known taxa See Workshop home page for more information (choose comparative genomics in drop down menu): http://workshop.molecularevolution.org/ecg/#wcgeur
Evolutionary Quantitative Genetics
03/09/2010
Start October 14 (1 meeting/week) AFLP: a tool box for studies of population genetic
03/09/2010
Course description
Researchers in the field of molecular ecology and evolution require versatile and low-cost genetic typing methods. The AFLP (amplified fragment length polymorphism) method was introduced 13 years ago and shows many features that fulfil these requirements. With good quality genomic DNA at hand, it is relatively easy to generate anonymous multilocus DNA profiles in most species and the start-up time before data can be generated is often less than a week. Built-in dynamic, yet simple modifications make it possible to find a protocol suitable to the genome size of the species and to screen thousands of loci in hundreds of individuals for a relatively low cost.
Conservation genetics in a changing world
03/09/2010
November 22-24
Fermentors, HPLC and Experimental Evolution
03/09/2010
Course description
Experimental evolution is an approach to capture the processes of mutation, drift and selection in action in the laboratory, and to run the evolutionary tape over and over again. We have set up an experimental facility that allows us to study how genes, populations and communities interact across the levels of biological organization to produce the patterns of genomic organization and change, adaptive radiation, biotic interactions and biodiversity patterns we can observe in nature. The critical equipment required for this approach is a large set of advanced fermentors for growing microorganisms (yeast and bacteria) in which experimental conditions can be carefully controlled and maintained during long periods of time, and analytical tools, such as HPLC, gas analyser and pulsed field gel electrophoresis.
Introduction to molecular ecology
03/09/2010
Course description
This is a practically oriented course providing the basics for DNA analyses in any kind of free living organism. Steps included are DNA extraction, PCR, DNA sequencing, primer design, microsatellite analyses, phylogenetic reconstructions and population genetic analyses of DNA sequences and polyallelic markers. Computer programs that will be used inlcude BioEdit, MEGA, DNAsp, FSTAT and STRUCTURE.